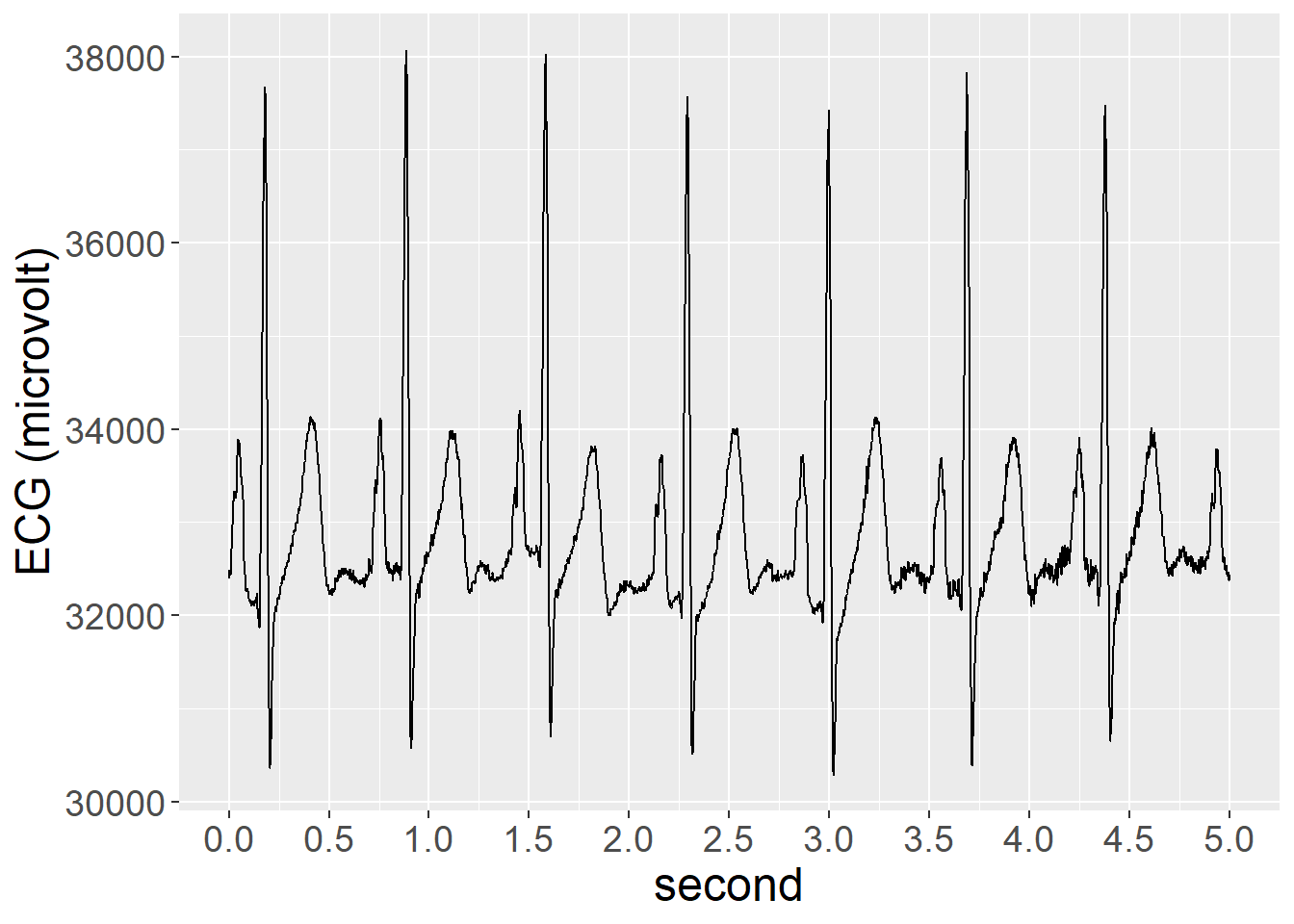
head(phy_raw_sample) row ID second ECG EDA
1 0 Pilot_003 0.000 32396 5743
2 1 Pilot_003 0.002 32456 5849
3 2 Pilot_003 0.004 32484 5791
4 3 Pilot_003 0.006 32452 5815
5 4 Pilot_003 0.008 32432 5775
6 5 Pilot_003 0.010 32530 5735
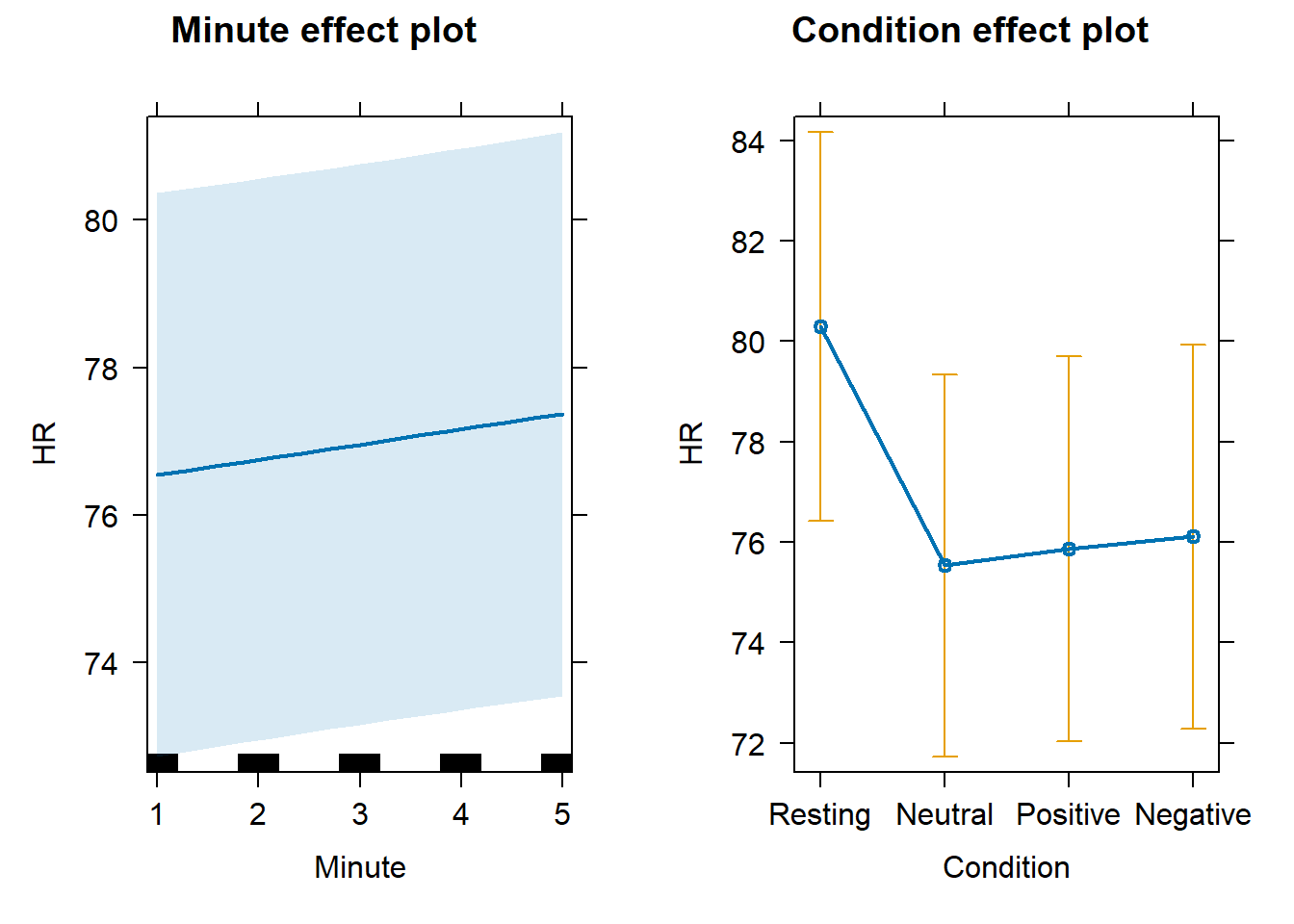
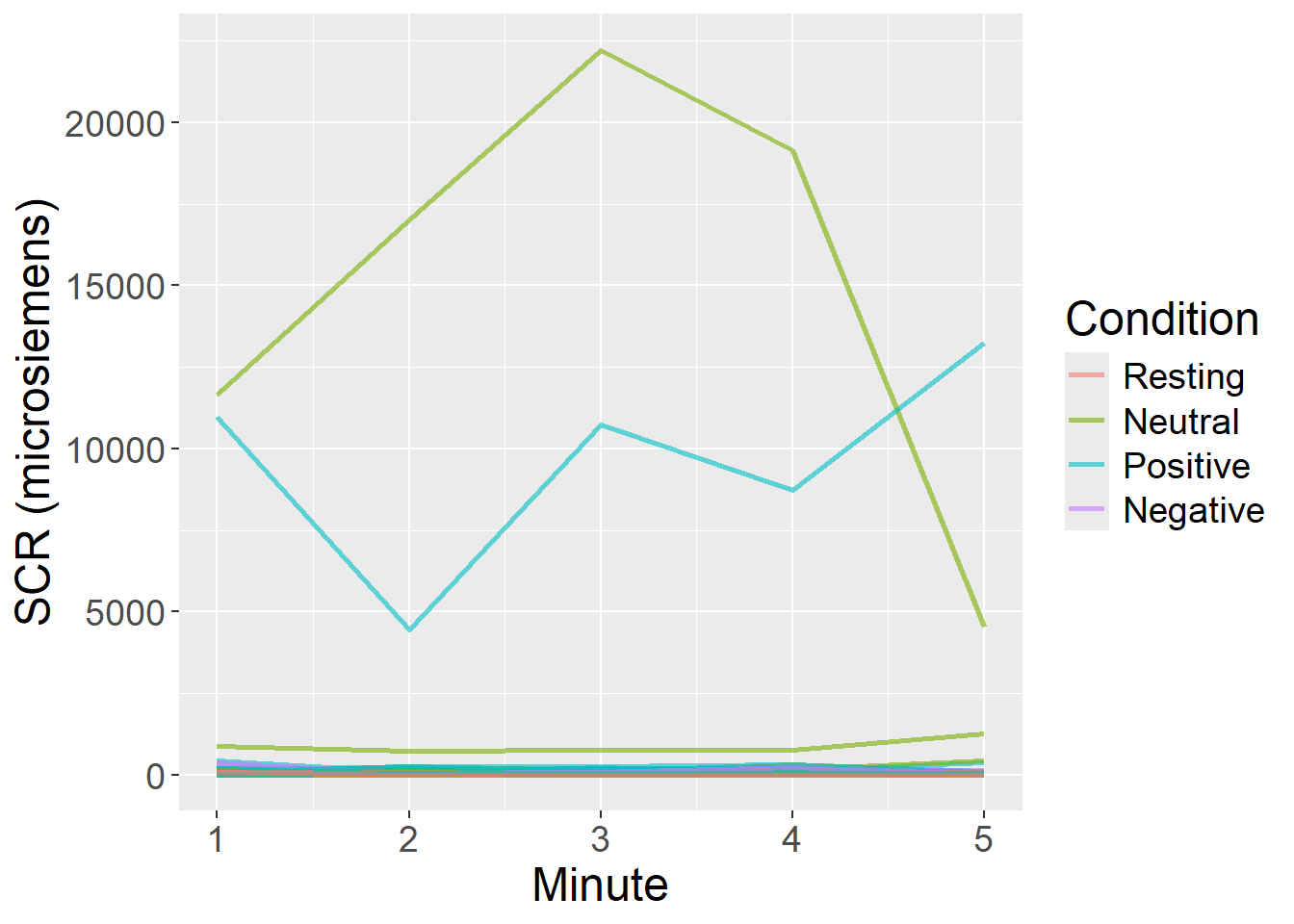
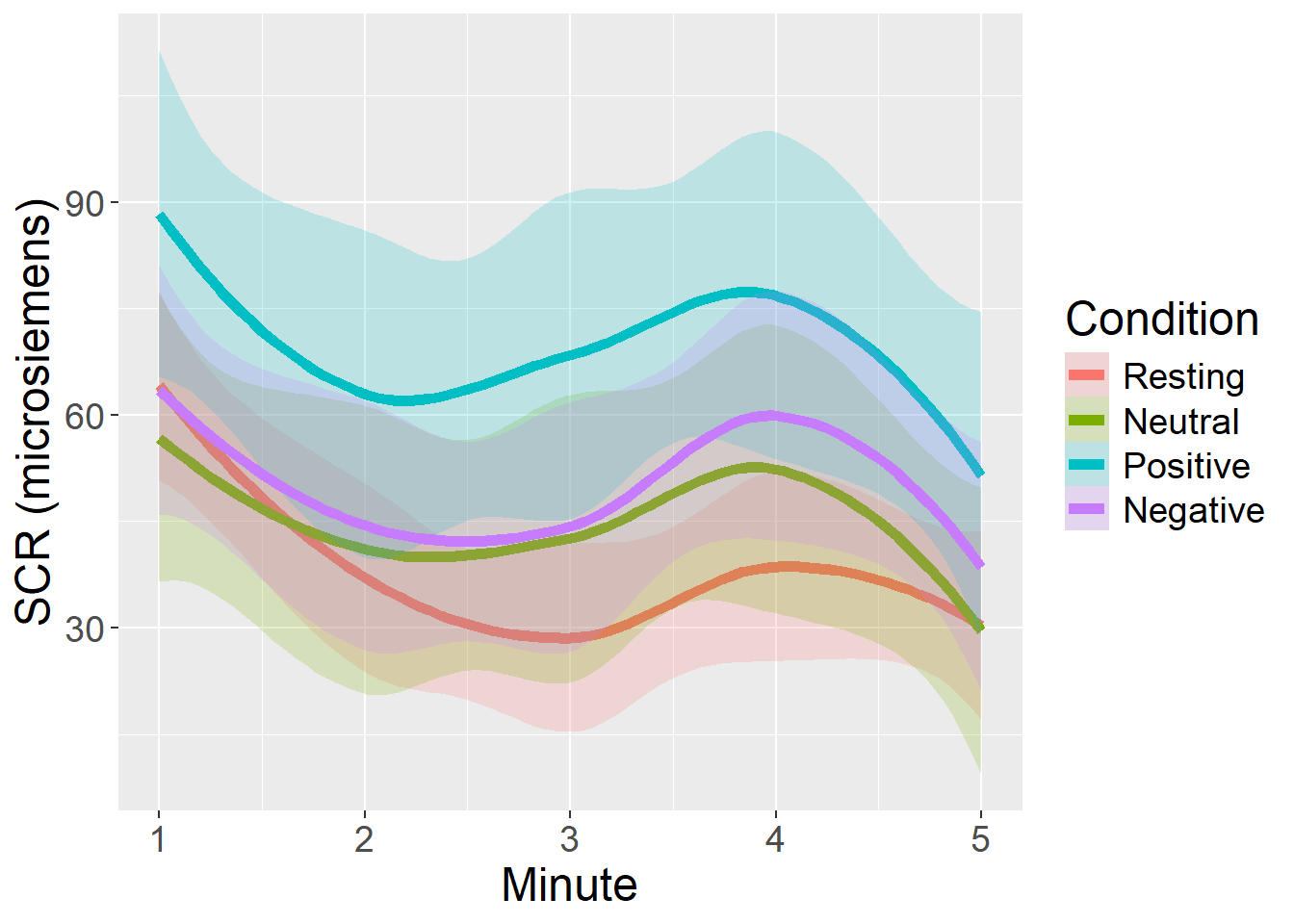
Comments and Conclusions
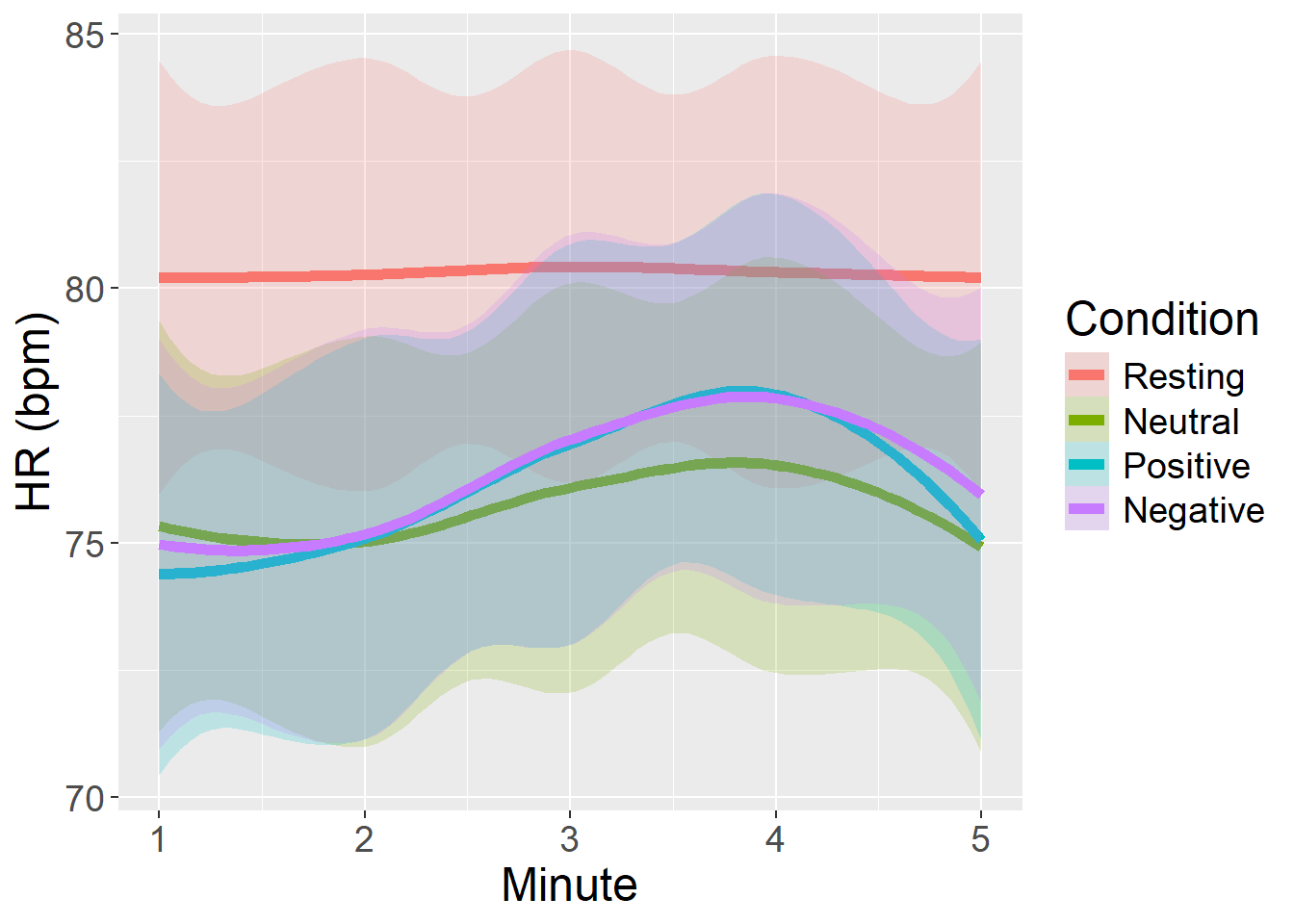
For Heart Rate, we observed highest mean values (and highest dispersion of scores) during the Resting state, while there were no relevant changes across the Neutral, Positive, or Negative video conditions, somehow unexpectedly. There was a slight tendency for HR to increase across time, independently from Condition.
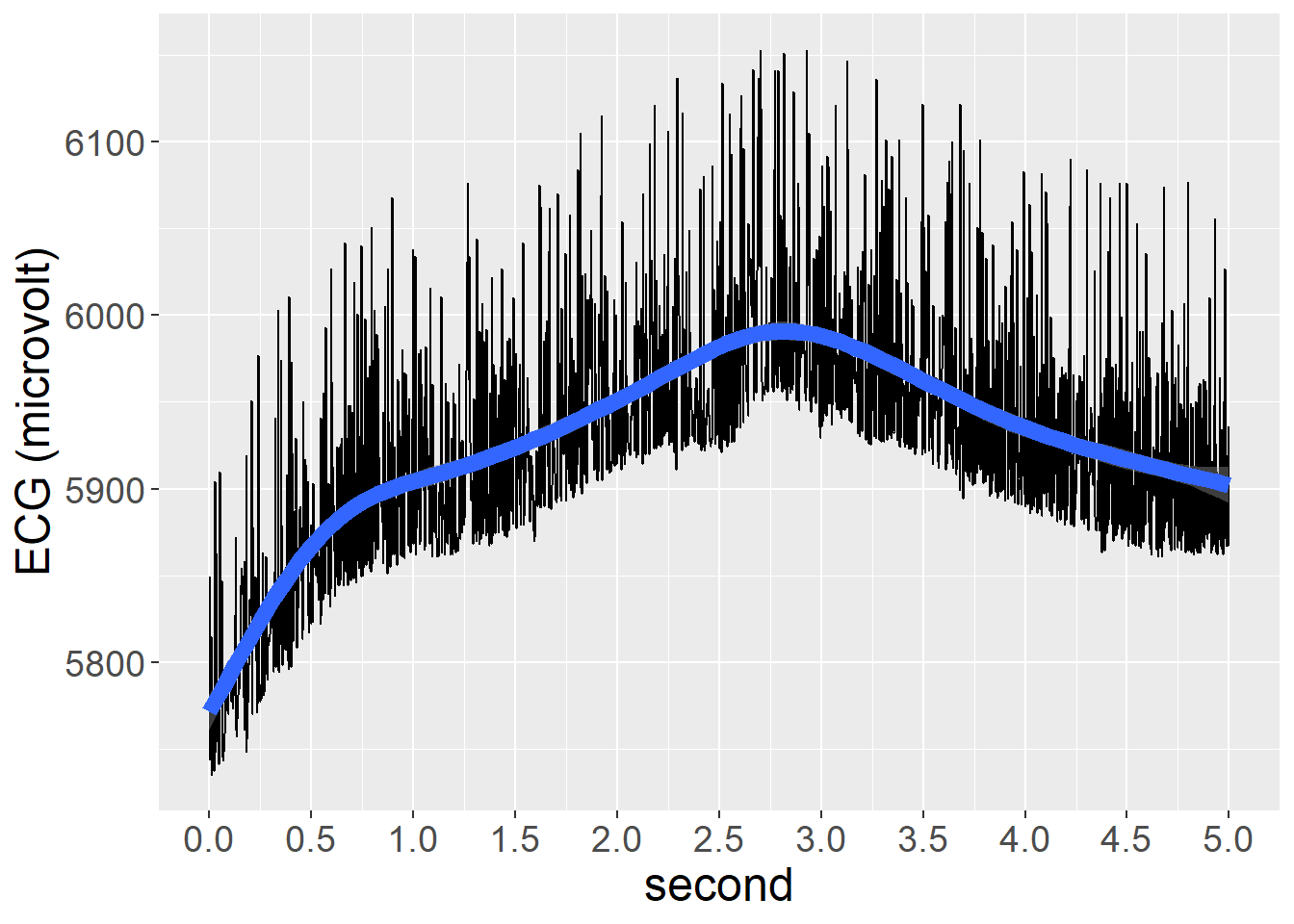
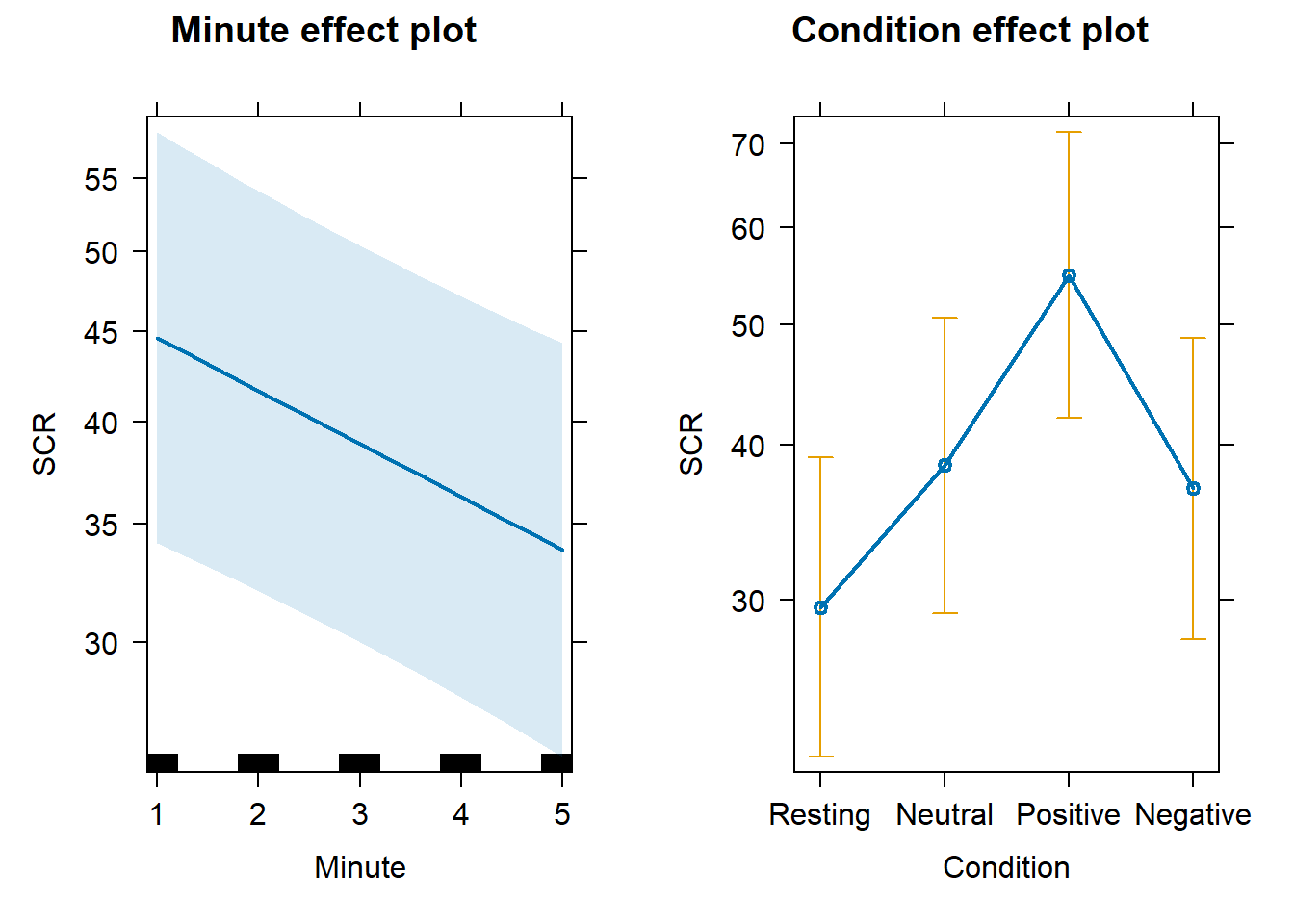
For Skin Conductance Response, we observed highest mean values during the Positive video condition, followed by Negative and Neutral conditions, with Resting state presenting the lowest values. Dispersion of scores also followed this patter. SCR also tended to reduce across time within each Condition, and quite independently from Condition.