Inspect Datasets
Let’s see the first few rows of each dataset for figuring out their structures. Note that Emotion varies within-subject
ID Order Emotion PrePost Trial ITI
1 4 NNP Positive Pre 1 993
2 4 NNP Positive Pre 2 206
3 4 NNP Positive Pre 3 171
4 4 NNP Positive Pre 4 154
5 4 NNP Positive Pre 5 169
6 4 NNP Positive Pre 6 169
ID Order Emotion Retro_Estimate Prosp_Estimate
1 4 NNP Positive 6.5 51089
2 4 NNP Neutral 5.0 85090
3 4 NNP Negative 4.0 65048
4 5 PNN Positive 7.0 56634
5 5 PNN Neutral 6.0 58890
6 5 PNN Negative 7.0 90949
Finger Tapping
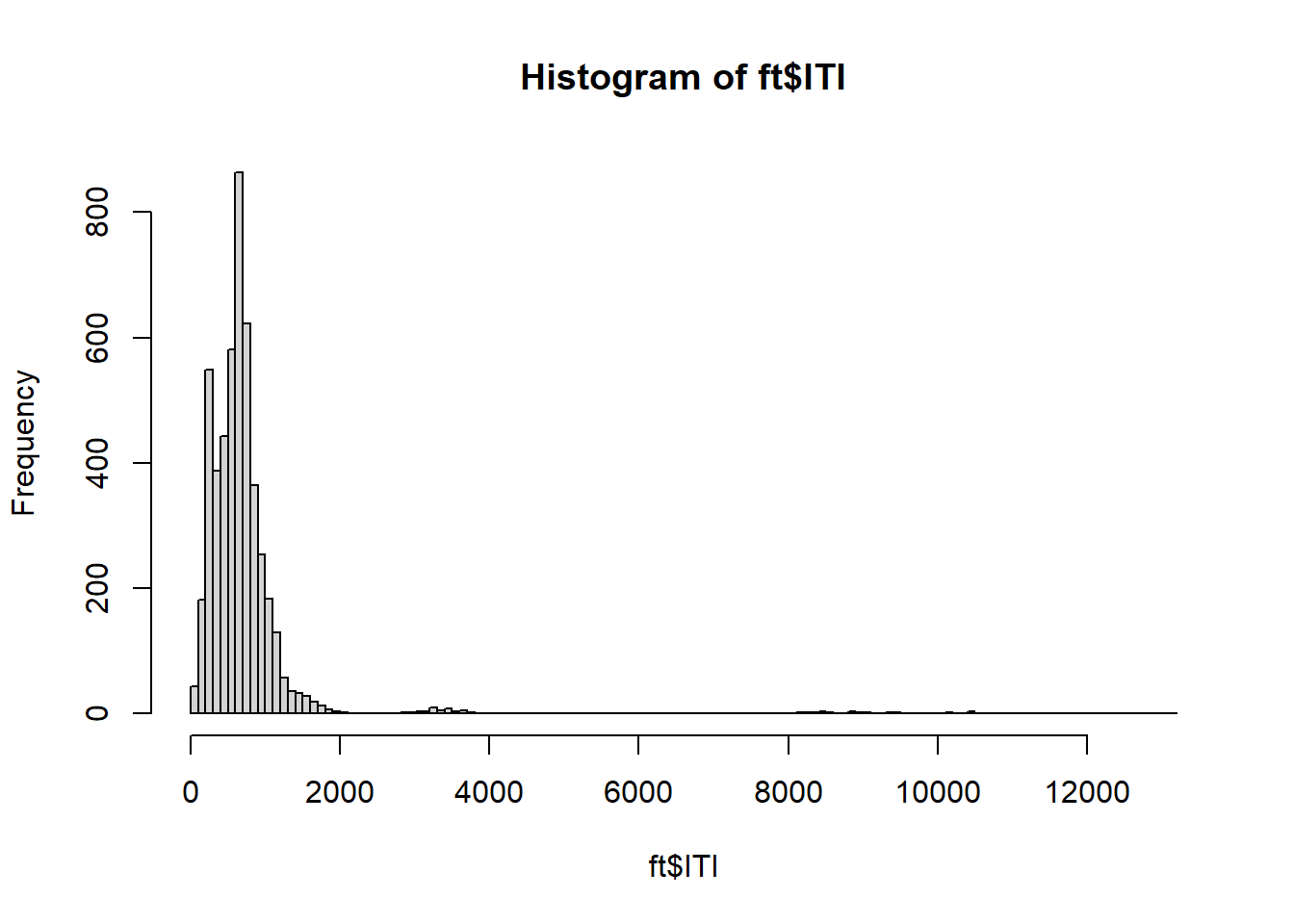
As recommended, let’s remove the first trial for each subject. Then, let’s inspect the distribution of ITI.
= ft[ft$ Trial != 1 , ]= ft[ft$ PrePost == "Post" , ]hist (ft$ ITI, breaks= 100 )
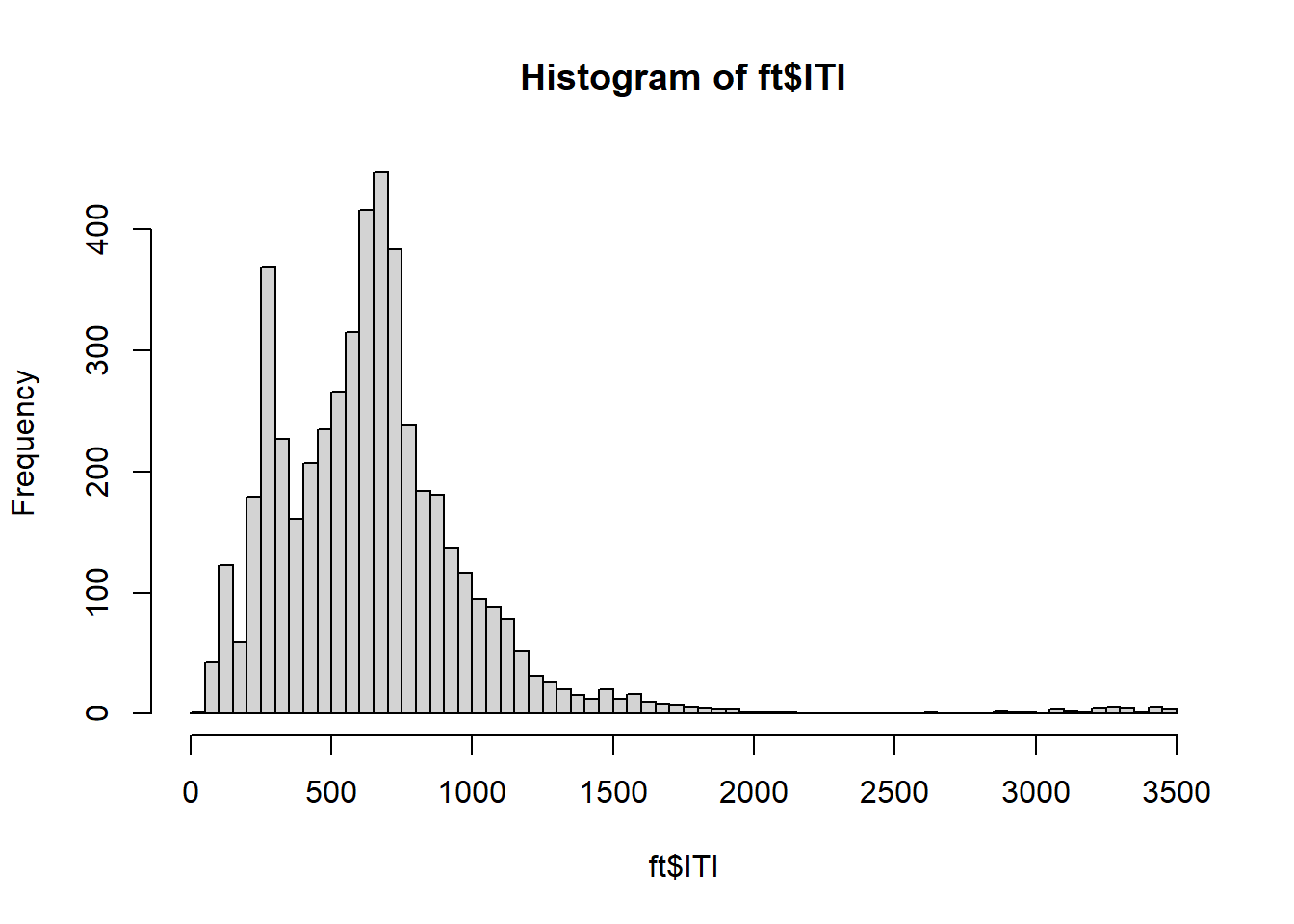
OUCH! There are a few outliers! Let’s remove ITI observations that are too far away from the average:
= (ft$ ITI - mean (ft$ ITI))/ sd (ft$ ITI)= ft[! abs (ITI_zScore) > 3 , ]hist (ft$ ITI, breaks= 100 )
Fit Statistical Model on Finger Tapping
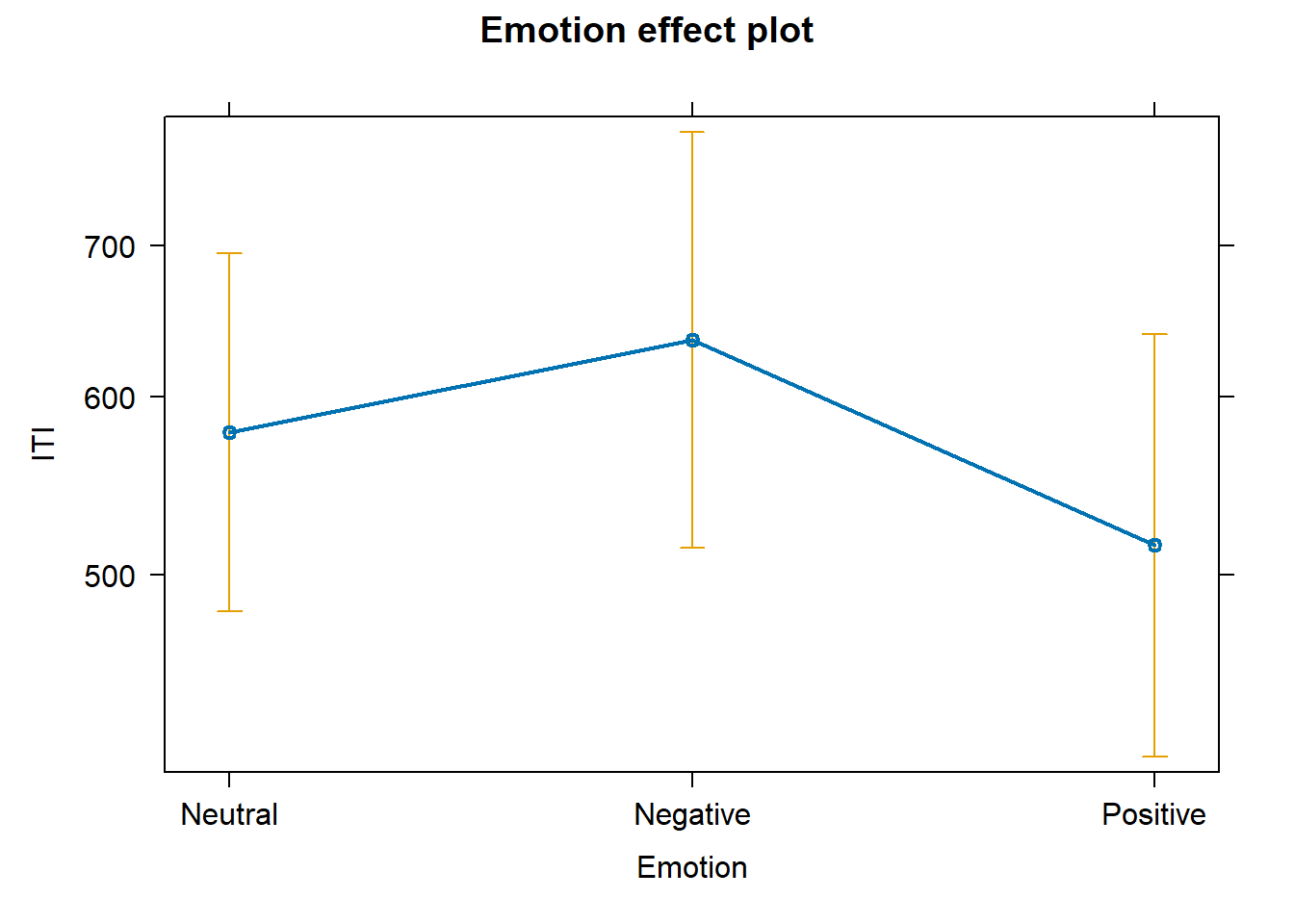
Because each participant provides multiple responses, we can test not only if Emotion affects average ITI, but also if it affects its dispersion. Let’s run a special model for this:
# prepare factorial variables $ ID = factor (ft$ ID)$ Emotion = factor (ft$ Emotion, levels= c ("Neutral" ,"Negative" ,"Positive" ))
# actually fit the model = glmmTMB (~ Emotion + (Emotion|| ID),dispformula = ~ Emotion ,data = ft,family = Gamma (link= "log" )
# see summary of model coefficients summary (fit_ft)
Family: Gamma ( log )
Formula: ITI ~ Emotion + (Emotion || ID)
Dispersion: ~Emotion
Data: ft
AIC BIC logLik deviance df.resid
54458.6 54516.9 -27220.3 54440.6 4820
Random effects:
Conditional model:
Groups Name Variance Std.Dev. Corr
ID (Intercept) 0.3202 0.5659
EmotionNegative 0.1089 0.3300 0.00
EmotionPositive 0.1275 0.3571 0.00 0.00
Number of obs: 4829, groups: ID, 37
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 6.36044 0.09307 68.34 <2e-16 ***
EmotionNegative 0.09439 0.05514 1.71 0.0869 .
EmotionPositive -0.11543 0.05884 -1.96 0.0498 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Dispersion model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 4.43535 0.03551 124.91 < 2e-16 ***
EmotionNegative -0.16648 0.05046 -3.30 0.00097 ***
EmotionPositive -0.04293 0.05013 -0.86 0.39181
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
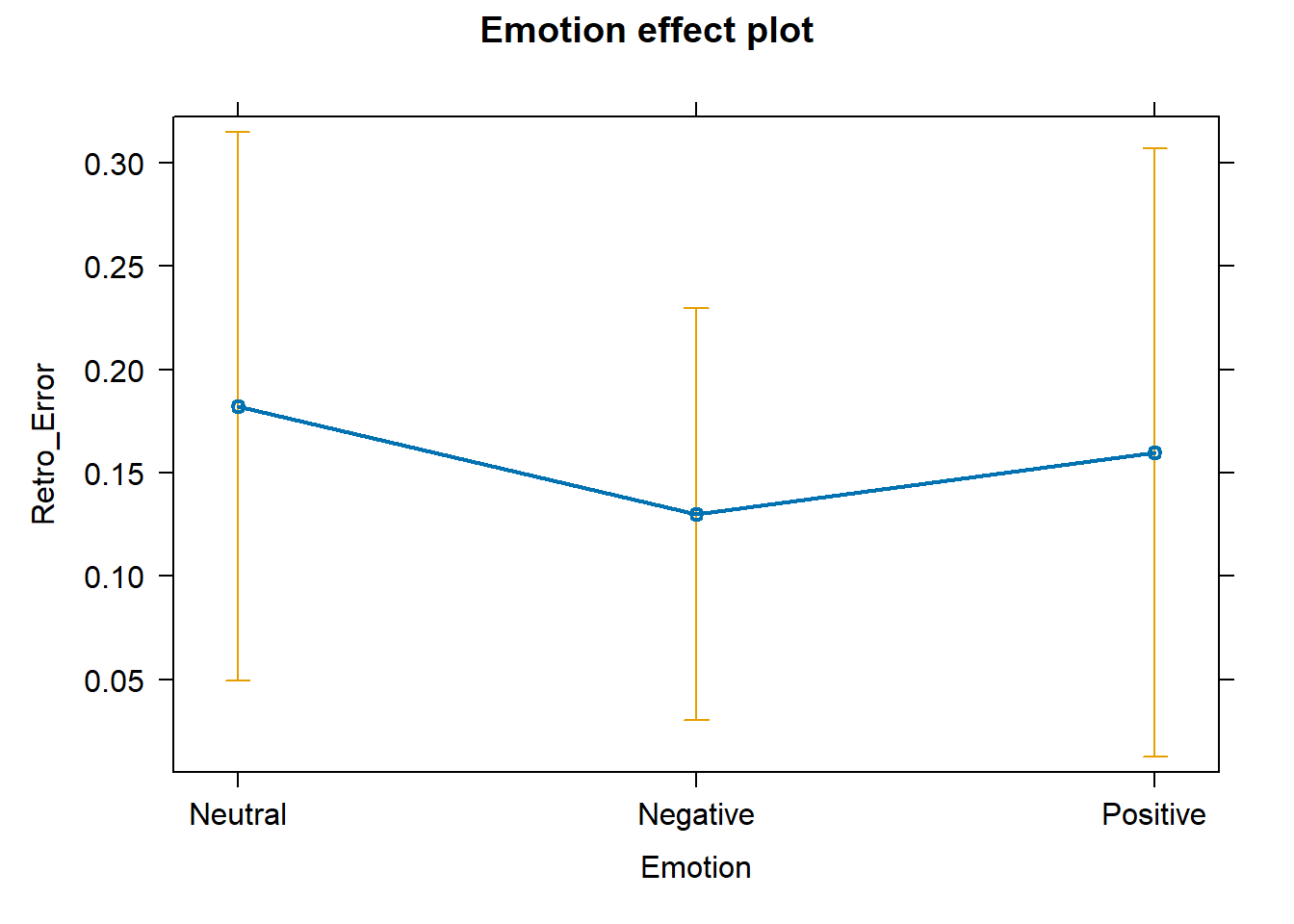
# plot predicted effects plot (allEffects (fit_ft))
Retrospective Time
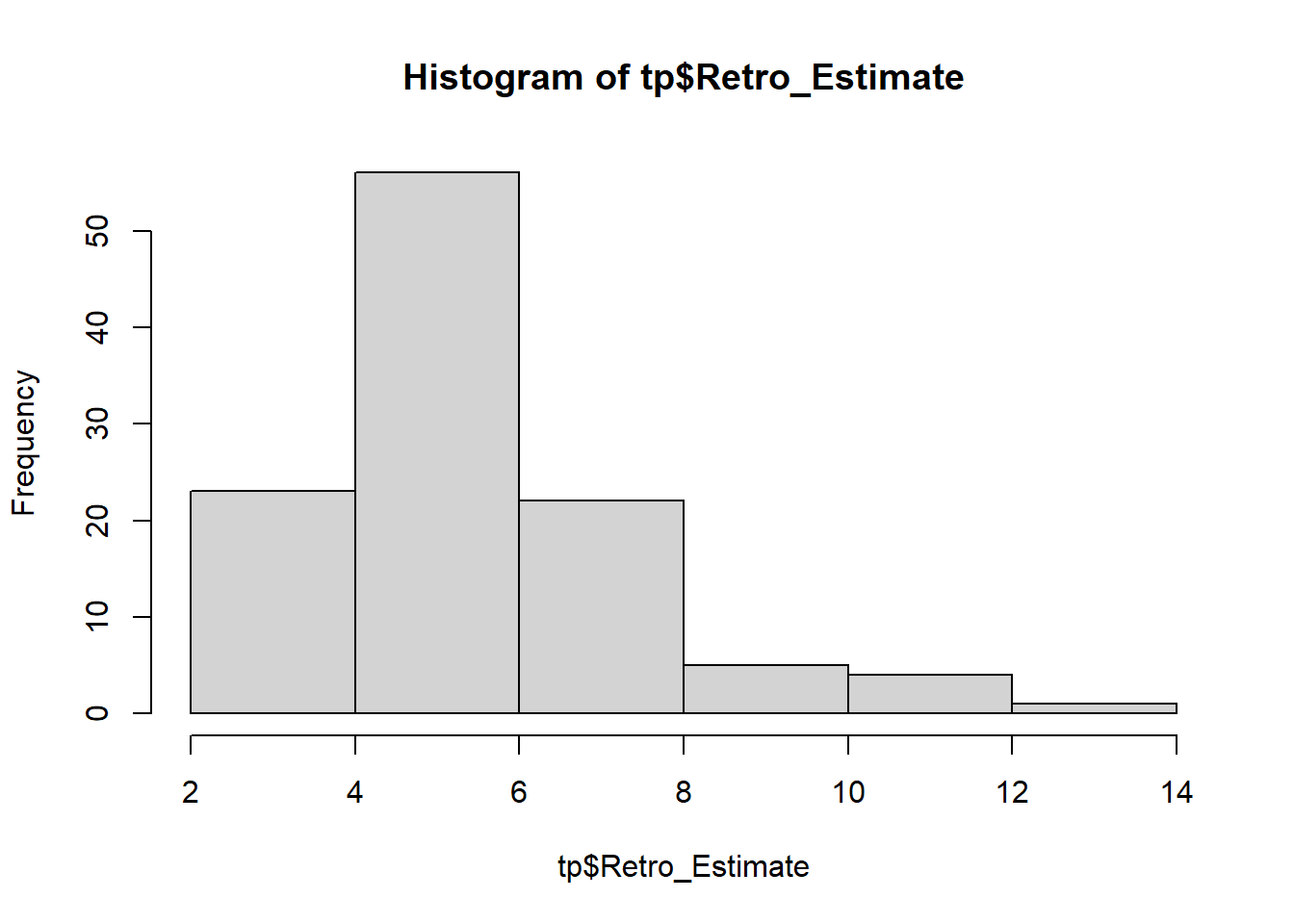
# inspect distribution of Retrospective estimates hist (tp$ Retro_Estimate)
Compute (in)accuracy as a ratio:
\[ RetrospectiveError = \frac{\text{Estimate} - \text{ObjectiveTime}}{\text{ObjectiveTime}} \]
# compute precision (error) of retrospective estimate $ Retro_Error = (tp$ Retro_Estimate - 5 ) / 5
⮩ Positive values indicate overestimation . Negative values indicate underestimation .
But… if a video is largely overestimated by some and largely underestimated by others, the average error is zero… does it mean perfect accuracy? No! Global accuracy vs inaccuracy is reflected by the total amount of dispersion average over- or under-estimation .
Fit Statistical Model on Retrospective estimates
# prepare factorial variables $ ID = factor (tp$ ID)$ Emotion = factor (tp$ Emotion, levels= c ("Neutral" ,"Negative" ,"Positive" ))
# actually fit the model = glmmTMB (~ Emotion + (1 | ID),dispformula = ~ Emotion,data = tp
# see summary of model coefficients summary (fit_re)
Family: gaussian ( identity )
Formula: Retro_Error ~ Emotion + (1 | ID)
Dispersion: ~Emotion
Data: tp
AIC BIC logLik deviance df.resid
73.2 92.2 -29.6 59.2 104
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
ID (Intercept) 0.08629 0.2938
Residual NA NA
Number of obs: 111, groups: ID, 37
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.18216 0.06689 2.723 0.00646 **
EmotionNegative -0.05243 0.04835 -1.084 0.27819
EmotionPositive -0.02243 0.07292 -0.308 0.75837
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Dispersion model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.5351 0.2562 -9.894 <2e-16 ***
EmotionNegative -2.3919 1.4955 -1.599 0.11
EmotionPositive 0.3938 0.3573 1.102 0.27
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
# plot predicted effects plot (allEffects (fit_re))
Prospective Time
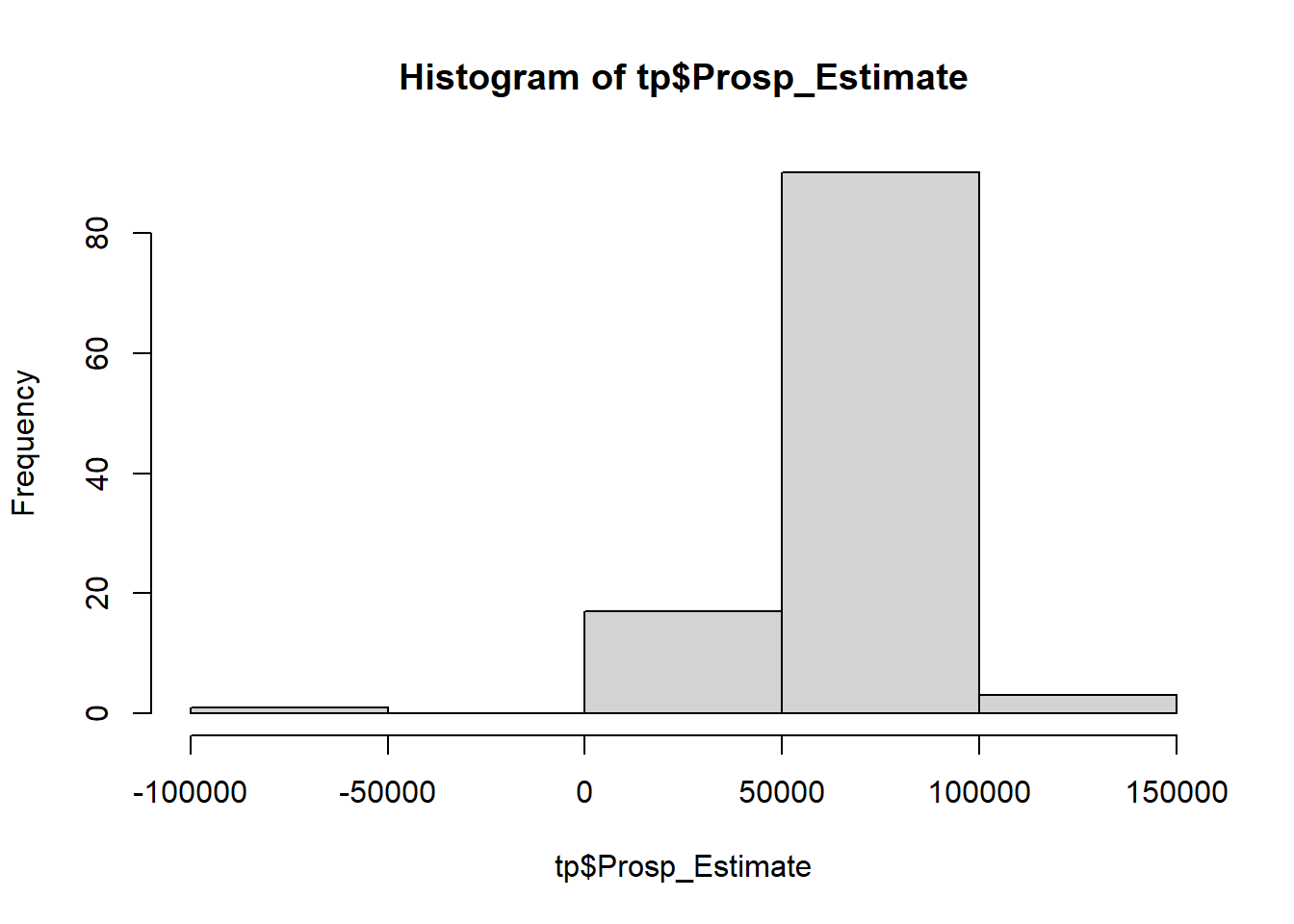
# inspect distribution of Prospective estimates hist (tp$ Prosp_Estimate)
OUCH! There is a negative value! Remove it:
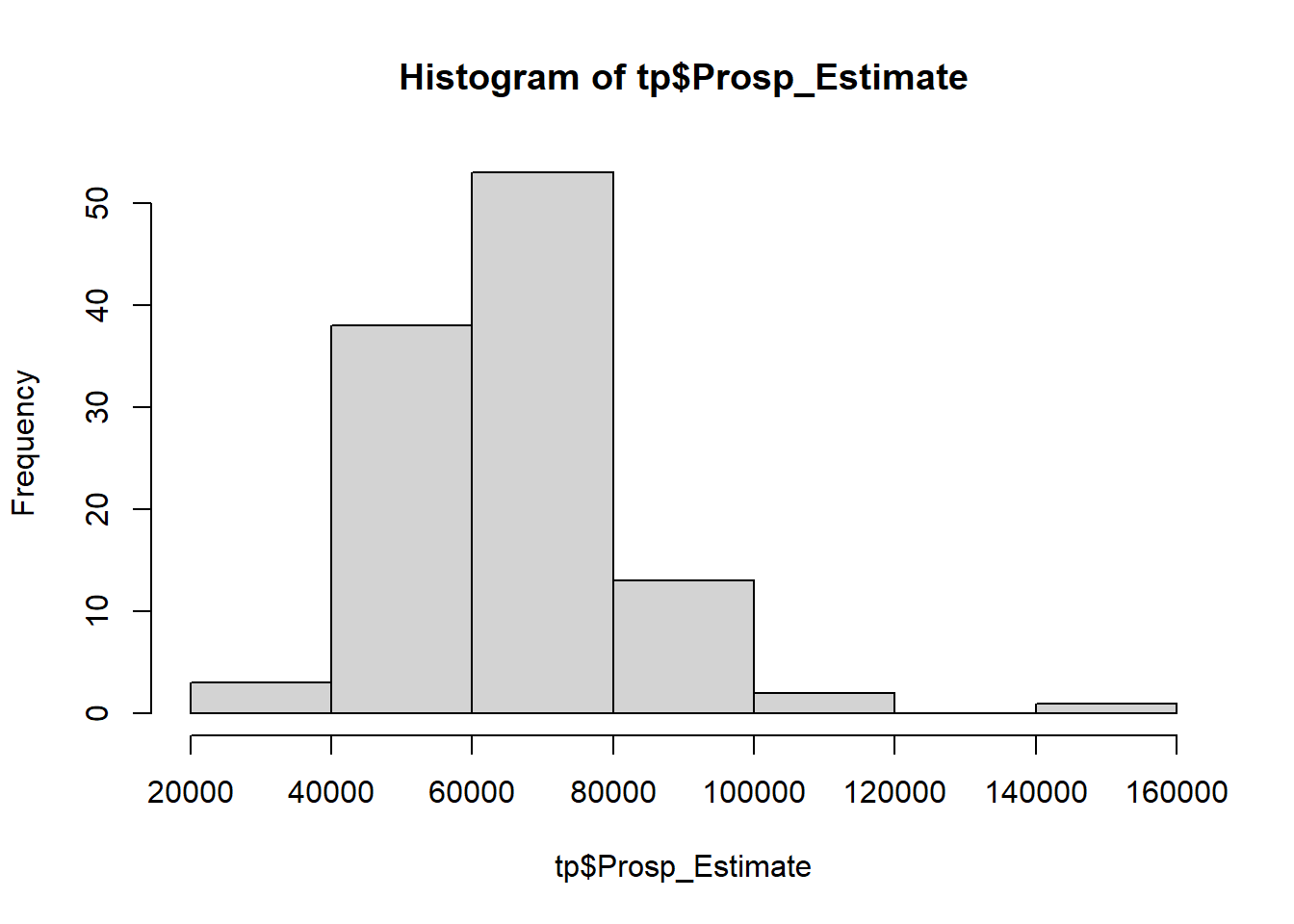
$ Prosp_Estimate[tp$ Prosp_Estimate < 0 ] = NA hist (tp$ Prosp_Estimate)
Compute error as a ratio:
\[ ProspectiveError = \frac{\text{Estimate} - \text{ObjectiveTime}}{\text{ObjectiveTime}} \]
# compute precision (error) of prospective estimate $ Prosp_Error = (tp$ Prosp_Estimate - 60000 ) / 60000
Fit Statistical Model on Prospective estimates
= glmmTMB (~ Emotion + (1 | ID),dispformula = ~ Emotion,data = tp
# see summary of model coefficients summary (fit_pro)
Family: gaussian ( identity )
Formula: Prosp_Error ~ Emotion + (1 | ID)
Dispersion: ~Emotion
Data: tp
AIC BIC logLik deviance df.resid
7.6 26.5 3.2 -6.4 103
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
ID (Intercept) 0.05073 0.2252
Residual NA NA
Number of obs: 110, groups: ID, 37
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.09613 0.04133 2.326 0.02002 *
EmotionNegative 0.11009 0.04019 2.739 0.00617 **
EmotionPositive -0.06066 0.03751 -1.617 0.10582
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Dispersion model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -4.3845 0.5722 -7.662 1.83e-14 ***
EmotionNegative 1.3005 0.6384 2.037 0.0417 *
EmotionPositive 1.1552 0.7290 1.585 0.1131
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
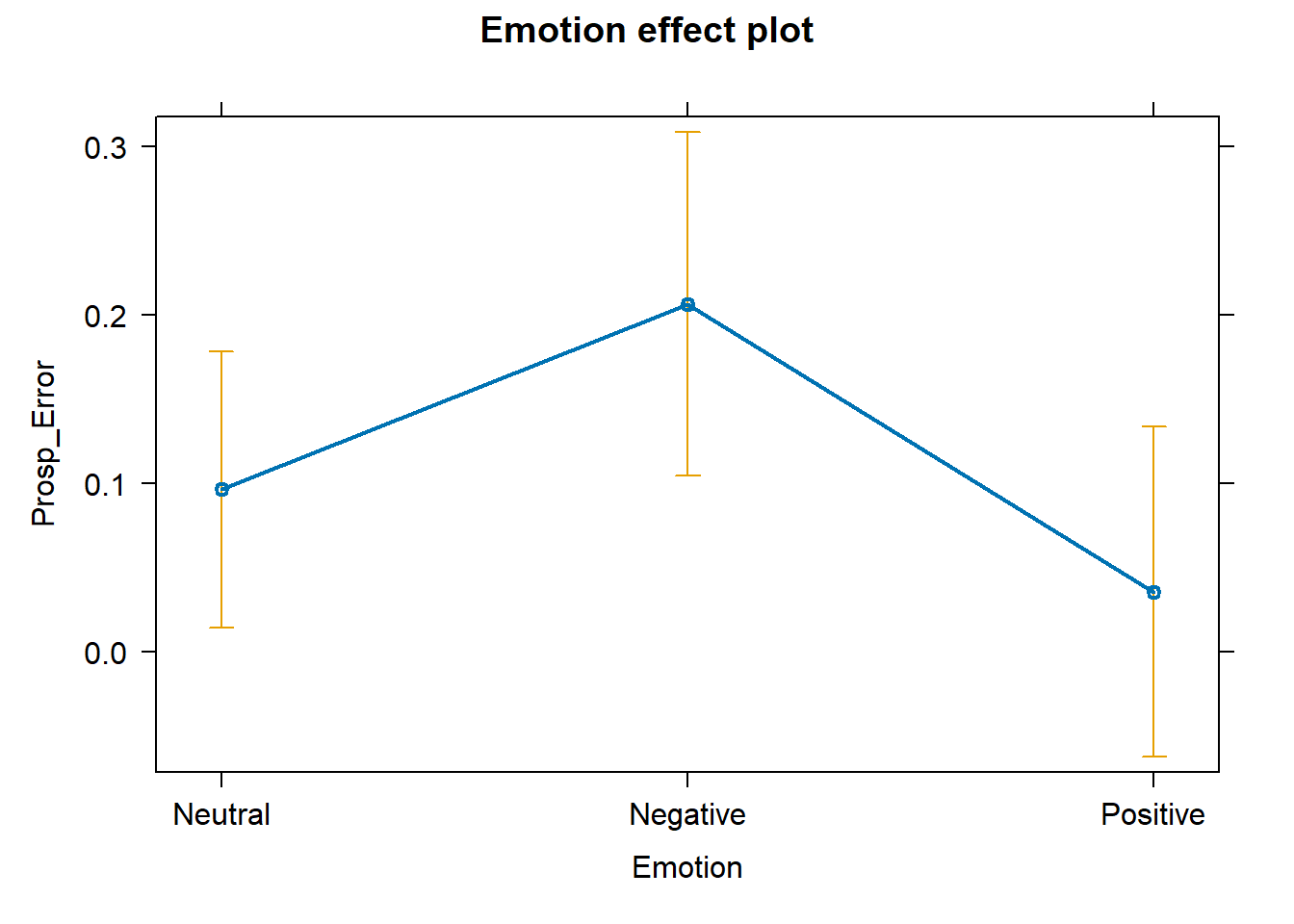
# plot predicted effects plot (allEffects (fit_pro))

Comments and Conclusions
An important point, when computing statistical analyses and finding relevant/significant effects, is to estimate how large they are. Since we used variables on interpretable metrics (ratio of error, milliseconds of ITI) the interpretation might be straightforward. However, standardized effect sizes (e.g., Cohen’s d) are also recommended.