import numpy as npBasics of Data Science:
Numpy and Pandas
Why NumPy and Pandas?
Most data science pipelines in Python rely on these two packages, as base Python lacks built-in data structures like vectors with vectorized operations, matrices / arrays, and data frames
numpy: efficient numerical computing (homogeneous arrays, vectorized operations) and many math, algebraic, and basic statistical functions;pandas: fundamental data structures for tabular data (DataFrames), with convenient indexing, reshaping, and data manipulation tools;
NumPy arrays
Remember the concept of “array” from Basics of R for Data Science?
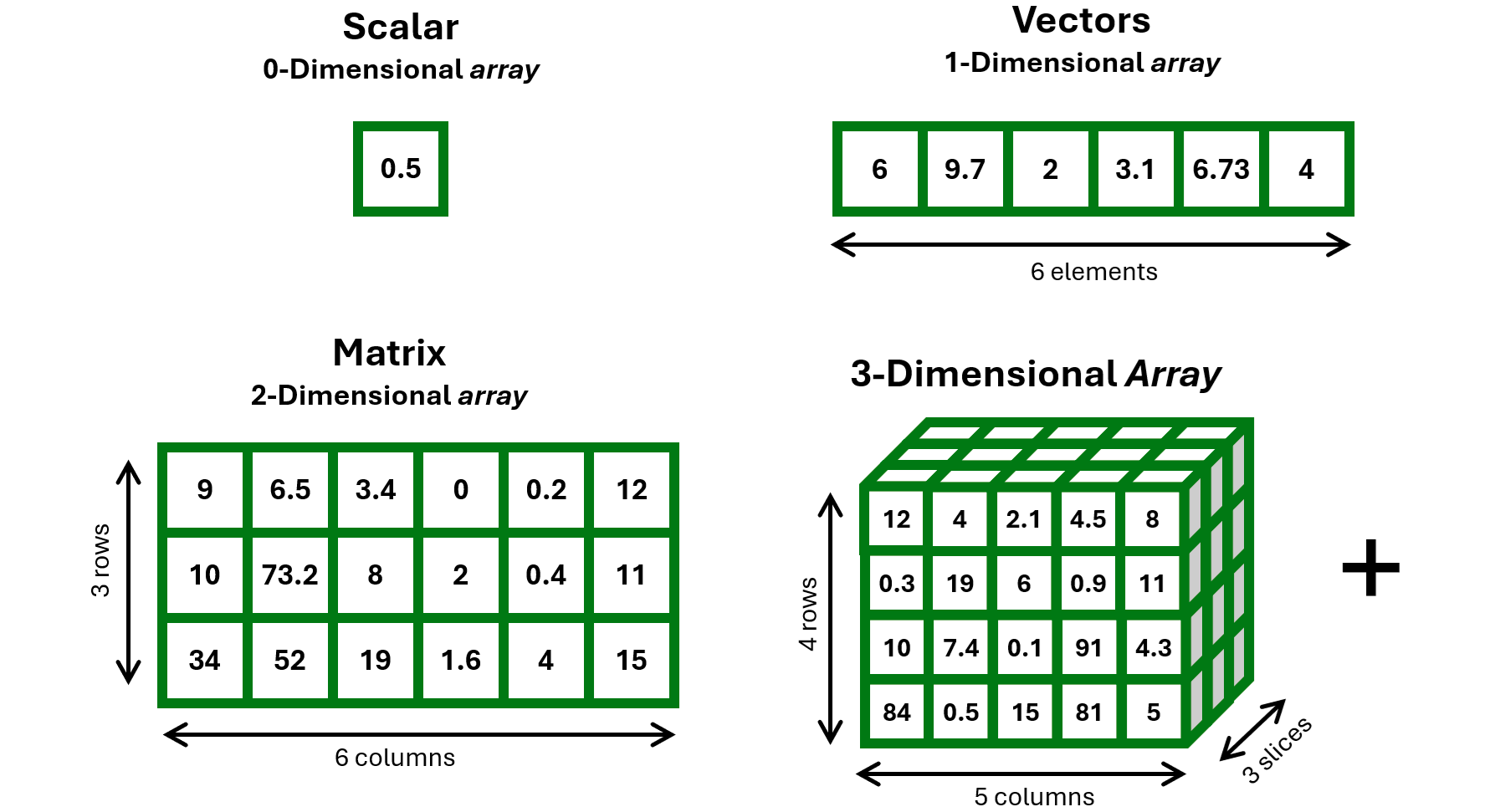
NumPy arrays
Example of 1-D array
Example of 2-D array (aka matrix, structured as a list of equally-long lists)
Arrays: Vectorized Operations
Unlike basic lists, arrays are homogenous and support fast vectorized operations, e.g., np.sqrt(x), x+5, x**2, etc.
array([[ 90., 65., 34., 0., 2., 120.],
[100., 732., 80., 20., 4., 110.],
[340., 520., 190., 16., 40., 150.]])Different ways of doing the same thing:
array([[3. , 2.5495, 1.8439, 0. , 0.4472, 3.4641],
[3.1623, 8.5557, 2.8284, 1.4142, 0.6325, 3.3166],
[5.831 , 7.2111, 4.3589, 1.2649, 2. , 3.873 ]])Arrays: Vectorized Operations
[ 6 8 10 12][ 5 12 21 32]70These features make np.array() conceptually very similar to the c() and array() objects of R. However, as we will see, np.array() is stricter in enforcing data types
Arrays: Structural attributes
What you can inspect for an array:
(3, 6)2188144dtype('float64')Arrays: Data types
dtype('float64')dtype('int64')dtype('<U2')dtype('<U12')dtype('<U21')Arrays: Data type coercion
Even with characters!
Arrays: Data type coercion
Actually, this “problem” is part of the secret of what makes Python numpy so efficient… but be careful!
How do you avoid this silent coercion? Explicitly state the data type from the beginning!
array([ 3.14159, 11. , 12. , 13. , 14. ])Arrays: Slicing and Indexing
np.int64(10)np.int64(50)array([20, 30, 40])Pandas Series
Series are essentially like 1D numpy arrays, but labelled with keys (numerical indexing is deprecated for Series)
Pandas DataFrame ❤️
pandas DataFrame is the classical tabular object we are familiar with; can be seen as a special type of Python dict (dictionary), composed of internally-homogeneous, equally-long, labelled lists
myDF = pd.DataFrame( {
"teacher": ["Pastore", "Granziol", "Feraco", "Altoe", "Marci"],
"course": ["CurrentIssues", "BasicsInference", "SEM", "Outliers", "QMP"],
"hours": [10, 20, 20, 5, 5],
"mandatory": [True, True, False, False, True],
"semester": [1, 1, 2, 2, 2]
} , index=["t1","t2","t3","t4","t5"])
myDF teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2DataFrame: Inspecting
(5, 5)Index(['teacher', 'course', 'hours', 'mandatory', 'semester'], dtype='object')<class 'pandas.core.frame.DataFrame'>
Index: 5 entries, t1 to t5
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 teacher 5 non-null object
1 course 5 non-null object
2 hours 5 non-null int64
3 mandatory 5 non-null bool
4 semester 5 non-null int64
dtypes: bool(1), int64(2), object(2)
memory usage: 205.0+ bytesteacher object
course object
hours int64
mandatory bool
semester int64
dtype: object hours semester
count 5.000000 5.000000
mean 12.000000 1.600000
std 7.582875 0.547723
min 5.000000 1.000000
25% 5.000000 1.000000
50% 10.000000 2.000000
75% 20.000000 2.000000
max 20.000000 2.000000about equivalent to str() and summary() in R
DataFrame: Indexing columns
teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2DataFrame: Indexing columns + row slicing
teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2DataFrame: Indexing multiple columns
teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2DataFrame: Indexing columns + row slicing
teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2DataFrame: Indexing rows
teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2DataFrame: Indexing multiple rows
teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2Access rows by position
DataFrame: Indexing rows + columns
teacher course hours mandatory semester
t1 Pastore CurrentIssues 10 True 1
t2 Granziol BasicsInference 20 True 1
t3 Feraco SEM 20 False 2
t4 Altoe Outliers 5 False 2
t5 Marci QMP 5 True 2
this is very similar to indexing using myDF[0,0] or myDF[0,"teacher"] in R
DataFrame: Indexing rows + columns
If you don’t like the row index, drop it!
teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1
1 Granziol BasicsInference 20 True 1
2 Feraco SEM 20 False 2
3 Altoe Outliers 5 False 2
4 Marci QMP 5 True 2
by default, rows have no names, and row names are not strictly needed in a DataFrame; without names, integer are used for indexing rows even using .loc[], which makes it more similar to R
DataFrame Logical indexing
teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1
1 Granziol BasicsInference 20 True 1
2 Feraco SEM 20 False 2
3 Altoe Outliers 5 False 2
4 Marci QMP 5 True 2 teacher course hours mandatory semester
1 Granziol BasicsInference 20 True 1
2 Feraco SEM 20 False 2 teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1 teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1
1 Granziol BasicsInference 20 True 1DataFrame Logical indexing
teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1
1 Granziol BasicsInference 20 True 1
2 Feraco SEM 20 False 2
3 Altoe Outliers 5 False 2
4 Marci QMP 5 True 2 teacher course
1 Granziol BasicsInference
2 Feraco SEM teacher course
0 Pastore CurrentIssues teacher course
0 Pastore CurrentIssues
1 Granziol BasicsInferenceDataFrame Logical indexing
teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1
1 Granziol BasicsInference 20 True 1
2 Feraco SEM 20 False 2
3 Altoe Outliers 5 False 2
4 Marci QMP 5 True 2 teacher course
1 Granziol BasicsInference
2 Feraco SEM teacher course
0 Pastore CurrentIssues teacher course
0 Pastore CurrentIssues
1 Granziol BasicsInferenceActually, this is the most recommended! especially for data manipulation
Add new Columns in a DataFrame
teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1
1 Granziol BasicsInference 20 True 1
2 Feraco SEM 20 False 2
3 Altoe Outliers 5 False 2
4 Marci QMP 5 True 2Add new Columns in a DataFrame
teacher course hours mandatory semester
0 Pastore CurrentIssues 10 True 1
1 Granziol BasicsInference 20 True 1
2 Feraco SEM 20 False 2
3 Altoe Outliers 5 False 2
4 Marci QMP 5 True 2Just index a column that doesn’t exist… and fill it!
Modify Columns in a DataFrame
teacher course hours mandatory semester wellBoh
0 Pastore CurrentIssues 10 True 1 48.711
1 Granziol BasicsInference 20 True 1 51.882
2 Feraco SEM 20 False 2 51.099
3 Altoe Outliers 5 False 2 49.029
4 Marci QMP 5 True 2 50.134DataFrame (Python pandas) vs data.frame (R)
| Task | Python (pandas) |
R |
|---|---|---|
| Select column | df["var"] |
df["var"] |
| Select column | df.var |
df$var |
| Multiple columns | df[["a", "b"]] |
df[, c("a", "b")] |
| Filter rows | df[df["var"] > 0] |
df[df$var > 0, ] |
| Add column | df["newVar"] = ... |
df$newVar = ... |
| Modify column | df["var"] = df["var"]+10 df["var"] += 10 |
df$var = df$var+10 |
| Summary | df.describe() |
summary(df) |
| Row access | df.loc[0] |
df[1, ] |
| Cell access | df.loc[0, "var"] |
df[1, "var"] |
| Cell access | df.iloc[0, 2] |
df[1, 3] |
Merging DataFrames (with pd.merge())
ID department age grade
0 Jane dpg 21.4 23
1 Jim dpg 22.5 28
2 John dpss 20.2 30
3 Jack dpss 23.0 15"name"! otherwise, add argument how="outer" to keep all values and possibly fill with NaN (missing values)
Concat. DataFrames (with pd.concat())
dfDPG = pd.DataFrame({
"name": ["Jane", "Jim"],
"grade": [28, 15],
"department": ["dpg", "dpg"]
})
dfDPG name grade department
0 Jane 28 dpg
1 Jim 15 dpgdfDPSS = pd.DataFrame({
"name": ["John", "Jack"],
"grade": [30, 23],
"department": ["dpss", "dpss"]
})
dfDPSS name grade department
0 John 30 dpss
1 Jack 23 dpssReshape Wide → Long (with .melt())
dfWide = pd.DataFrame({
"ID": ["id01", "id02", "id03"],
"gender": ["m", "m", "f"],
"T0": [-2.31, -1.70, -2.64],
"T1": [-1.50, -0.86, -2.30]
})
dfWide ID gender T0 T1
0 id01 m -2.31 -1.50
1 id02 m -1.70 -0.86
2 id03 f -2.64 -2.30dfLong = dfWide.melt(id_vars=["ID","gender"], var_name="TIME",
value_name="SCORE", value_vars=["T0","T1"])
dfLong ID gender TIME SCORE
0 id01 m T0 -2.31
1 id02 m T0 -1.70
2 id03 f T0 -2.64
3 id01 m T1 -1.50
4 id02 m T1 -0.86
5 id03 f T1 -2.30Reshape Long → Wide (with .pivot())
dfLong = pd.DataFrame({
"ID": ["id01", "id01", "id02", "id02", "id03", "id03"],
"gender": ["m", "m", "m", "m", "f", "f"],
"TIME": ["T0", "T1", "T0", "T1", "T0", "T1"],
"SCORE": [-2.31, -1.50, -1.70, -0.86, -2.64, -2.30]
}); dfLong ID gender TIME SCORE
0 id01 m T0 -2.31
1 id01 m T1 -1.50
2 id02 m T0 -1.70
3 id02 m T1 -0.86
4 id03 f T0 -2.64
5 id03 f T1 -2.30TIME ID gender T0 T1
0 id01 m -2.31 -1.50
1 id02 m -1.70 -0.86
2 id03 f -2.64 -2.30Grouped operations (with .groupby())
teacher course hours mandatory semester wellBoh
0 Pastore CurrentIssues 10 True 1 48.711
1 Granziol BasicsInference 20 True 1 100051.882
2 Feraco SEM 20 False 2 100051.099
3 Altoe Outliers 5 False 2 49.029
4 Marci QMP 5 True 2 50.134Grouped operations (with .groupby())
Compute statistics for multiple columns by group
Apply multiple functions with .agg()
hours wellBoh
mean 12.00 40050.17
std 7.58 54773.46
min 5.00 48.71
max 20.00 100051.88myDF.groupby("semester").agg({"hours": ["mean","std","min","max"],
"wellBoh": ["mean","std","min","max"]
}).round(2) hours wellBoh
mean std min max mean std min max
semester
1 15.0 7.07 10 20 50050.30 70712.92 48.71 100051.88
2 10.0 8.66 5 20 33383.42 57735.90 49.03 100051.10