python -m venv nameOfMyEnvVirtual Environments, Packages, Import/Export
Virtual Environment
– an isolated Python environment that allows you to install packages and manage dependencies separately from your system/main Python installation. This helps avoid conflicts between projects that may require different versions of the same packages. venvs are routinely used in professional projects. We will not use them systematically in this course, but know that they are best practice for managing projects ensuring reproducibility
BTW, something similar now exists also in R via the renv package, although unfortunately not widely used
Virtual Environment
Practically, it is a local folder with an isolated full Python environment. It contains:
- a copy of the Python interpreter;
- all the packages you install at the exact versions used at that time
This folder can ideally be placed inside your project directory
Virtual Environment
Create a virtual environment with this command in your bash/terminal:
then activate it before using
…alternatively, inside IDEs, you may activate the venv via specific commands like reticulate::use_virtualenv("nameOfMyEnv", required=T) (in RStudio), or setting the Python interpreter manually and then restarting the kernel (in Spyder)
Virtual Environment
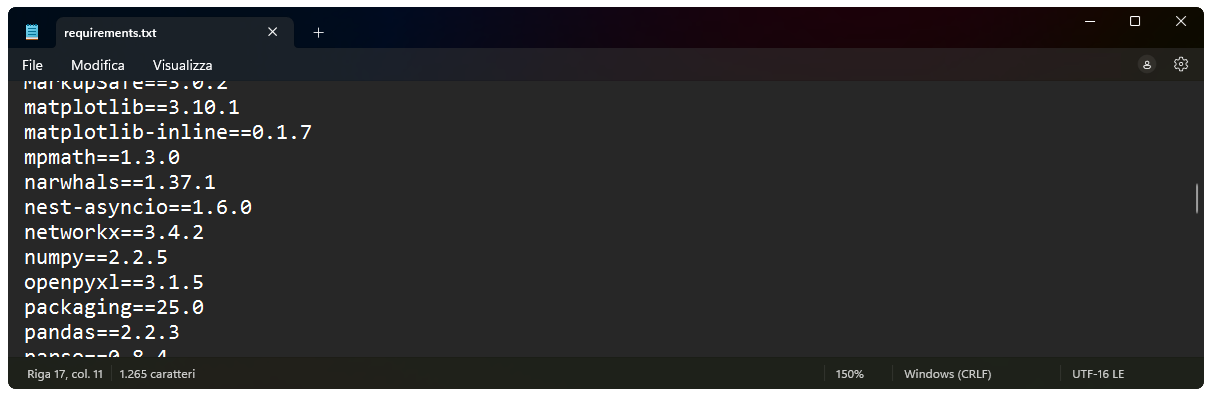
Regardless of your main Python installation, you should reinstall all packages needed for your project inside the local venv (after activating it). This is considered best practice, as it ensures isolation across projects and reproducibility. At any time, you can export a requirements.txt file to document the exact versions of all installed packages (this is particularly useful for sharing your environment, e.g., via GitHub):
Project like a pro 😎
- Use a virtual environment per project (as a subfolder);
- Place scripts, notebooks, and data in different subfolders;
- Use relative paths (e.g.,
"data/myfile.csv"); - Save results and figures via code (not manually)
myProjectFolder/
├── venv/ ← virtual environment
├── data/ ← .csv, .xlsx, etc.
├── scripts/ ← .py scripts
├── results/ ← output files, figures
├── notebooks/ ← markdowns, colab notebooks, etc.
├── requirements.txt ← list of installed packages for reproducibility
└── README.md ← brief description of the projectInstalling and importing packages
Installing, inside an IDE console or Colab:
Then, before using any of their functions, import packages and modules:
as gives a shorter alias to a package or module name (e.g., pd for pandas, np for numpy); this is convenient because in Python you frequently need to call different functions by always specifying the package/module name (unlike in R; unless you import individual functions, e.g., from numpy import array)
Using functions, help, autocomplete
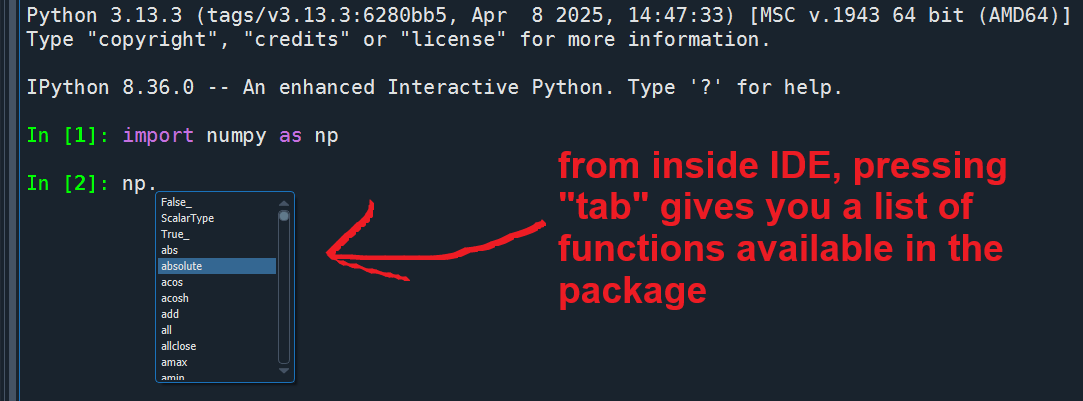
Use a function from a package, and call help:
Use tab to autocomplete and explore available functions of a package ↴
Using functions, help, autocomplete
As in R, you can rely on positional order of arguments instead of naming them, or you can completely omit them if there are valid default arguments. However, it’s best practice to make all relevant arguments explicit for readability and reproducibility
Accessing functions as methods
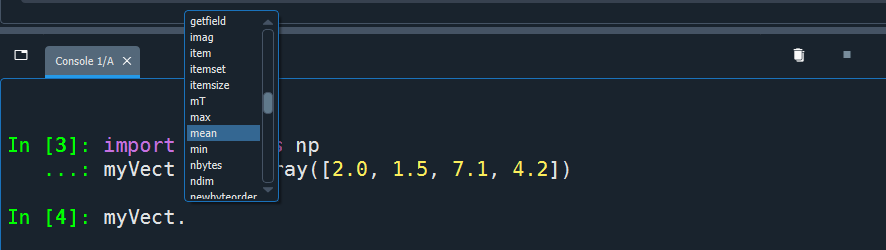
In Python, objects may have functions attached to them: these are called methods, and are accessed using dot (.) notation (more on this later!)
Use tab to autocomplete and explore available methods of an object →
Working directory
getwd() / setwd():
(in Colab, paths are relative to the notebook location in Google Drive)
Import/Export objects (no equivalent of save.image() of R)
simpler version
(this simpler version is suboptimal because it doesn’t properly close the file after using, but still works)
Import tabular data (more on pandas later!)
from CSV
from Excel
from Ctrl+C copied elements (beautiful ❤️ but only for Windows)
Export tabular data
Export figures
Delete an object (similar to rm(df) in R)
Listing all objects in workspace (similar to ls() in R)
dir()
dir() is a built-in function that does more than just returning a list of objects in workspace; it allows you to inspect all attributes and methods of any object
['append', 'clear', 'copy', 'count', 'extend', 'index', 'insert', 'pop', 'remove', 'reverse']['all', 'any', 'argmax', 'argmin', 'argpartition', 'argsort', 'astype', 'base', 'byteswap', 'choose', 'clip', 'compress', 'conj', 'conjugate', 'copy', 'ctypes', 'cumprod', 'cumsum', 'data', 'device', 'diagonal', 'dot', 'dtype', 'dump', 'dumps', 'fill', 'flags', 'flat', 'flatten', 'getfield']['abs', 'absolute', 'acos', 'acosh', 'add', 'all', 'allclose', 'amax', 'amin', 'angle', 'any', 'append', 'apply_along_axis', 'apply_over_axes', 'arange', 'arccos', 'arccosh', 'arcsin', 'arcsinh', 'arctan', 'arctan2', 'arctanh', 'argmax', 'argmin', 'argpartition', 'argsort', 'argwhere', 'around', 'array', 'array2string']